Targeted Proteome Assays
|
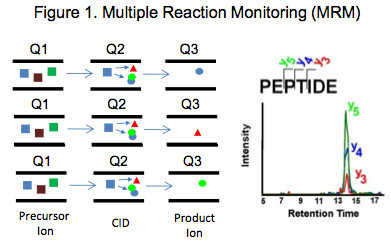
Targeted Proteomics was just recently selected as Nature Method of the Year for 2012 and is recognized as the most sensitive and specific mass spectrometric analysis method to detect and quantitate pre-selected components in a complex sample matrix such as a proteolytic digest of a plasma or tissue extract [1-3]. Multiple Reaction Monitoring (MRM) which is one of the most commonly used targeted approaches available, utilizes a triple quadrupole mass spectrometer carrying out sequential rounds of filtered precursor-product ion pair detection to concurrently quantitate multiple analytes as schematically depicted in
Figure 1. Utilizing MRM we have built Targeted Proteome Assays (TPAs), which are 90 minute quantitative scheduled LC-MRM analyses for determining relative concentrations of about 50-200 pre-selected protein biomarkers of interest in several selected tissues. Some of the targeted proteins are sufficiently common that they may also be quantified in other complex biological samples and tissue types[4, 5].
|
 |
||||||||||||
|
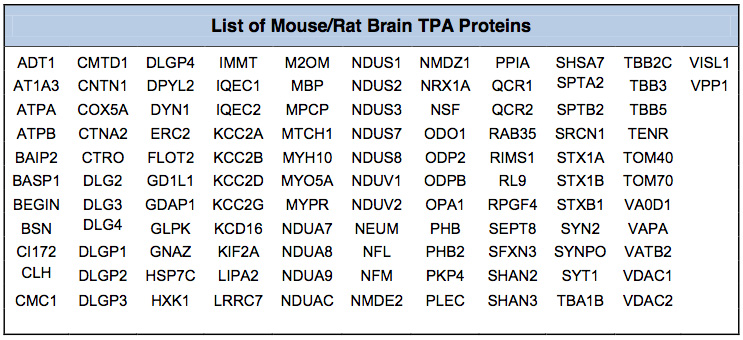
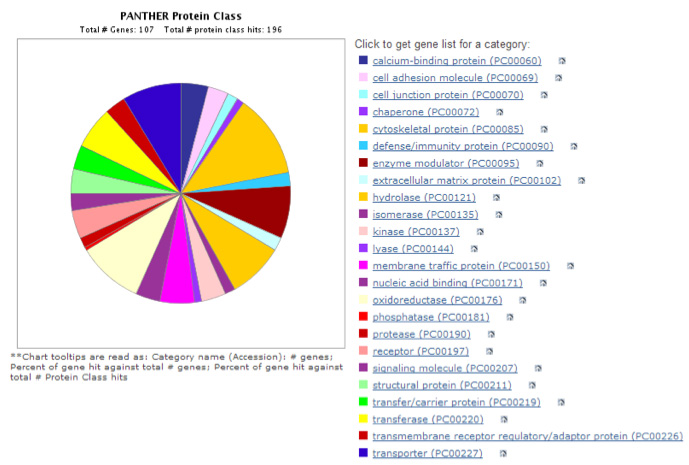
Mouse/Rat Brain Assay (MBA/112 Proteins): The Mouse/Rat Brain Proteome Assay interrogates the relative level of expression of 112 proteins (Table below) from 24 different protein classes (Figure 2). The assay was developed and optimized for Rat Cortex Post-synaptic density (PSD) preparations. We have also utilized this assay for Cortex Synaptoneurosomes and Striatal PSD. When this assay is used on other brain fractions the number of quantified proteins might be somewhat less than 112.
|
|||||||||||||
|
|||||||||||||
|
|||||||||||||
|
Assay Details: Targeted Proteome Assays (TPAs) include tryptic digestion, C18 clean-up, and 90 min LC-MRM runs on an AB SCIEX 5500 QTRAP MS (Figure 3). Quantitation of each targeted protein is based on up to 15 data points (3 peptides x 5 transitions/peptide). A minimum of 10ug protein/sample is needed. We inject 1ug of protein digest in a single injection and for triplicate injections we run the technical replicates in a block randomized order. Subsequent data analysis includes:
(https://medicine.yale.edu/keck/proteomics/servicesfees/yped/) |

|
||||||||||||
|
|||||||||||||